An open-source R package for omics data integrative analysis in terms of
network, evolution and ontology
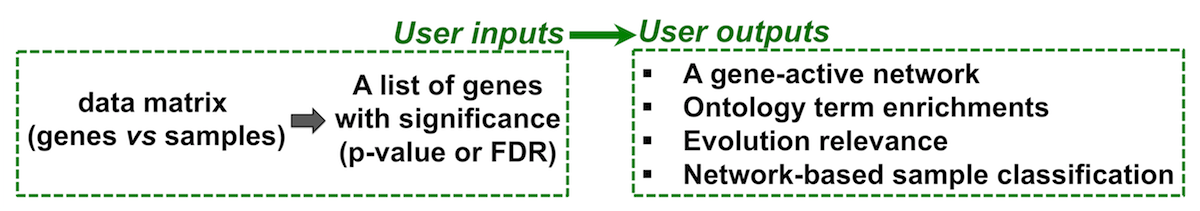
User I/O
Features
- Identification of gene-active networks from high-throughput omics data (e.g. mutation, expression, and replication timing);
- Network-based sample classifications and visualisations on 2D sample landscape;
- Random Walk with Restart for network affinity calculation;
- Semantic similarity between ontology terms (and between their annotated genes);
- Enrichment analysis using a variety of built-in databases;
- A wide variety of built-in RData: ontologies (including
Gene Ontology,Disease Ontology,Human PhenotypeandMammalian Phenotype), gene evolutionary age information and gene association networks in well-studied organisms (including human, mouse, rat, chicken, c.elegans, fruitfly, zebrafish and arabidopsis); - Support for high-performance parallel computing;
- A tool with rich visuals but less inputs, which can run on
Windows,MacandLinux.
More
Manual
Demos
- Demo for TCGA mutation and survival dataset
- Demo for multilayer replication timing dataset
- Demo for patient expression dataset
- Demo for time-course expression dataset
URL
Dependency
- Depends: igraph, supraHex
- Imports: graph, Rgraphviz, Matrix
- Suggests: limma, survival, foreach, doParallel, Biobase
- Extends: